Abstract
Introduction: The epigenome of T-cell acute lymphoblastic leukemia (T-ALL) is characterized by DNA hypermethylation at CpG islands and altered chromatin accessibility. However, it remains unclear when and how these changes of the epigenome contribute to the development of T-ALL.
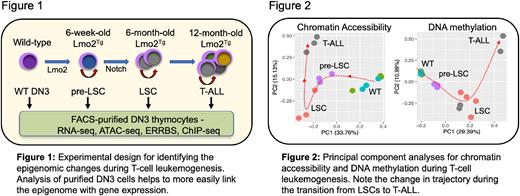
Methods: To better understand their role in T-ALL, we have examined the epigenome at three stages of T-cell leukemogenesis using the Lmo2 transgenic (Lmo2Tg) mouse model of human early thymocyte progenitor (ETP)-ALL (Fig. 1).
1. Pre-leukemic stem cells (pre-LSCs) isolated from 6-week-old mice, in which double negative 3 (DN3) thymocytes have long-term self-renewal enabling the development of T-ALL but with a long latency of 6 months.
2. Leukemic stem cells (LSCs) from 6-month-old mice, in which DN3 thymocytes have acquired the additional (epi) genetic mutations required for rapidly generating T-ALL when transplanted into recipient mice.
3. DN3 cells from 10 to 12-month-old mice that have succumbed to proliferative T-ALL.
DN3 cells from each of these stages of T-cell leukemogenesis together with age-matched wild-type controls were FACS-purified for gene expression (RNA-seq), chromatin accessibility (Assay of Transposase Accessible Chromatin sequencing, ATAC-seq), DNA methylation (Enhanced Reduced Representation Bisulfite Sequencing, ERRBS). Abnormalities in the epigenome directly related to Lmo2 binding were investigated by integration with CHIP-seq.
Results: Overall, principal component analyses demonstrated distinct trajectories for chromatin, DNA methylation and gene expression changes during leukemogenesis (Fig. 2).
In pre-LSCs, chromatin was more accessible in CpG open seas enriched for the heptad transcription factor (TF) binding sites of hematopoietic stem cells (Runx1, Fli1, Tal1, Lyl1, Gata2, Lmo2, Erg), and less accessible in CpG islands enriched for Klf15 binding sites. Lmo2 directly bound to 1,831 more accessible chromatin regions and was associated with gene activation of the stem cell heptad TFs. In addition to this 'stem-cell’ chromatin pattern, pre-LSCs had DNA hypermethylation in CpG open seas enriched for Runx1 and Fli1 binding. There was also the beginning of the more classic DNA hypermethylation of GpG islands reported for human T-ALL but this was not associated with gene repression. Instead, GpG islands with increased heterogeneity of DNA methylation had closed chromatin and associated gene repression (R=-0.16, p=0.01).
In LSCs, chromatin changes remained relatively stable, with increased accessibility at heptad TF binding regions and decreased accessibility in Klf15 binding sites. In contrast, there was a marked increase in DNA hypermethylation of bivalent promoters that was strongly associated with gene repression of several putative tumour suppressors including Klf4 and Tet2 (R=-0.31, p<0.01). Importantly, many of these hypermethylated regions (41%) in LSCs were preceded by increased heterogeneity in pre-LSCs.
The most striking observations were those in the transition from LSCs to proliferative T-ALL, where there was reversal of many of the preceding chromatin changes, including closure of the stem cell heptad TF binding sites and re-opening at Klf15 binding in CpG islands. These changes were associated with gene expression changes characterised by loss of the stem cell program and increase in T-cell differentiation. While DNA hypermethylation at promoters was maintained, there was new DNA hypomethylation in lamina-associated late replicating regions.
Conclusion: This temporal analysis of purified cell populations facilitates integration of epigenomic changes with gene expression, which is a powerful approach to address the 'chicken-or-egg’ dilemma of the cancer epigenome. Our studies of the epigenome during T-cell leukemogenesis reveal a far more dynamic process than previously recognized and impossible to discern by analysis of T-ALL alone. Specifically, heterogeneity of DNA methylation precedes DNA hypermethylation at bivalent promoters while DNA hypomethylation at late replicating domains occurs much later. In contrast, we show that the T-cell oncogene establishes an early stem cell-like chromatin structure that is reversed in overt T-ALL. We postulate that DNA hypermethylation and gene repression of tumour suppressors in LSCs allows a 'relaxation’ of the chromatin to enable additional adaptive changes required for the rapid growth of T-ALL.
Disclosures
Wong:Pacific Edge Limited: Consultancy, Current equity holder in publicly-traded company.
Author notes
Asterisk with author names denotes non-ASH members.